The four-parameter Weibull functions
W4.Rd'W1.4' and 'W2.4' provide the four-parameter Weibull functions, self starter function and names of the parameters.
W1.4(fixed = c(NA, NA, NA, NA), names = c("b", "c", "d", "e"), ...) W2.4(fixed = c(NA, NA, NA, NA), names = c("b", "c", "d", "e"), ...)
Arguments
| fixed | numeric vector. Specifies which parameters are fixed and at what value they are fixed. NAs for parameter that are not fixed. |
|---|---|
| names | a vector of character strings giving the names of the parameters. The default is reasonable. |
| ... | additional arguments to be passed from the convenience functions. |
Details
The equations for the mean functions are given at weibull1.
Value
See weibull1.
References
Seber, G. A. F. and Wild, C. J (1989) Nonlinear Regression, New York: Wiley \& Sons (pp. 330--331).
Ritz, C (2009) Towards a unified approach to dose-response modeling in ecotoxicology To appear in Environ Toxicol Chem.
Note
This function is for use with the model fitting function drm.
See also
Setting \(c=0\) yields W1.3. A more flexible function, allowing
fixing or constraining parameters, is available through weibull1.
Examples
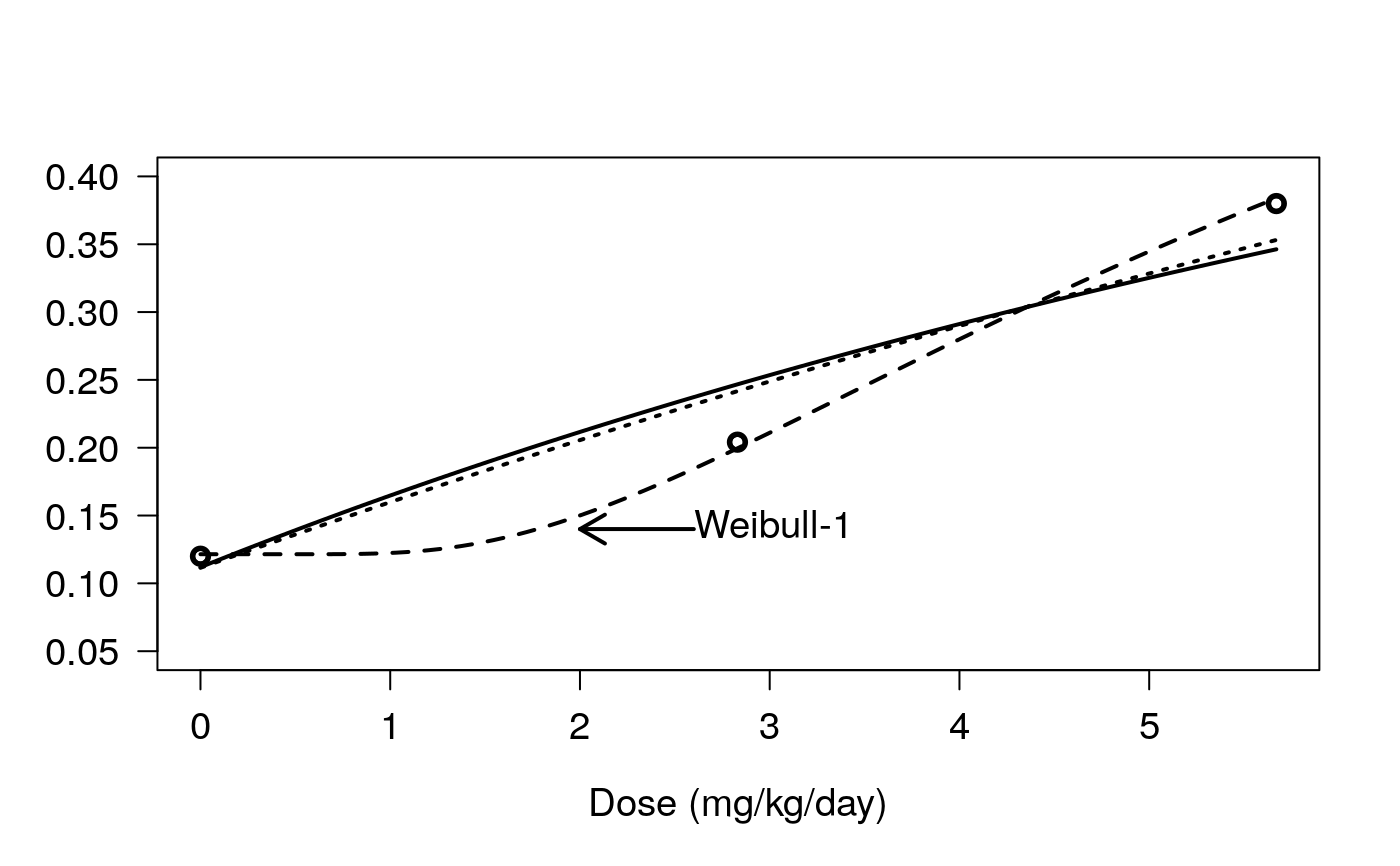
## Fitting a four-parameter Weibull (type 1) model terbuthylazin.m1 <- drm(rgr~dose, data = terbuthylazin, fct = W1.4()) summary(terbuthylazin.m1)#> #> Model fitted: Weibull (type 1) (4 parms) #> #> Parameter estimates: #> #> Estimate Std. Error t-value p-value #> b:(Intercept) 9.7401e-01 1.4451e-01 6.7401 3.753e-07 *** #> c:(Intercept) 2.4632e-02 1.4782e-02 1.6663 0.1076 #> d:(Intercept) 3.0889e-01 9.5904e-03 32.2080 < 2.2e-16 *** #> e:(Intercept) 2.5485e+02 4.0604e+01 6.2766 1.211e-06 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Residual standard error: #> #> 0.02694557 (26 degrees of freedom)## Fitting a first-order multistage model ## to data from BMDS by EPA ## (Figure 3 in Ritz (2009)) bmds.ex1 <- data.frame(ad.dose=c(0,50,100), dose=c(0, 2.83, 5.67), num=c(6,10,19), total=c(50,49,50)) bmds.ex1.m1<-drm(num/total~dose, weights=total, data=bmds.ex1, fct=W2.4(fixed=c(1,NA,1,NA)), type="binomial") modelFit(bmds.ex1.m1) # same as in BMDS#> Goodness-of-fit test #> #> Df Chisq value p value #> #> DRC model 1 0.57224 0.4494summary(bmds.ex1.m1) # same background estimate as in BMDS#> #> Model fitted: Weibull (type 2) (2 parms) #> #> Parameter estimates: #> #> Estimate Std. Error t-value p-value #> c:(Intercept) 0.111475 0.041719 2.6721 0.007539 ** #> e:(Intercept) 17.860515 5.919727 3.0171 0.002552 ** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1logLik(bmds.ex1.m1)#> 'log Lik.' -6.179627 (df=2)## BMD estimate identical to BMDS result ## BMDL estimate differs from BMDS result (different method) ED(bmds.ex1.m1, 10, ci="delta")#> #> Estimated effective doses #> #> Estimate Std. Error #> e:1:10 1.88179 0.62371## Better fit bmds.ex1.m2<-drm(num/total~dose, weights=total, data=bmds.ex1, fct=W1.4(fixed=c(-1,NA,1,NA)), type="binomial") modelFit(bmds.ex1.m2)#> Goodness-of-fit test #> #> Df Chisq value p value #> #> DRC model 1 0.010589 0.918summary(bmds.ex1.m2)#> #> Model fitted: Weibull (type 1) (2 parms) #> #> Parameter estimates: #> #> Estimate Std. Error t-value p-value #> c:(Intercept) 0.121468 0.044268 2.7439 0.006071 ** #> e:(Intercept) 6.850273 1.540986 4.4454 8.774e-06 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1#> #> Estimated effective doses #> #> Estimate Std. Error #> e:1:50 9.8829 2.2232## Creating Figure 3 in Ritz (2009) bmds.ex1.m3 <- drm(num/total~dose, weights=total, data=bmds.ex1, fct=LL.4(fixed=c(-1,NA,1,NA)), type="binomial") plot(bmds.ex1.m1, ylim = c(0.05, 0.4), log = "", lty = 3, lwd = 2, xlab = "Dose (mg/kg/day)", ylab = "", cex=1.2, cex.axis=1.2, cex.lab=1.2)mtext("Tumor incidence", 2, line=4, cex=1.2) # tailored y axis labelplot(bmds.ex1.m2, ylim = c(0.05, 0.4), log = "", add = TRUE, lty = 2, lwd = 2)plot(bmds.ex1.m3, ylim = c(0.05, 0.4), log = "", add = TRUE, lty = 1, lwd = 2)arrows(2.6 , 0.14, 2, 0.14, 0.15, lwd=2)text(2.5, 0.14, "Weibull-1", pos=4, cex=1.2)