Asymptotic regression model
AR.RdProviding the mean function and the corresponding self starter function for the asymptotic regression model.
AR.2(fixed = c(NA, NA), names = c("d", "e"), ...) AR.3(fixed = c(NA, NA, NA), names = c("c", "d", "e"), ...)
Arguments
| fixed | numeric vector. Specifies which parameters are fixed and at what value they are fixed. NAs for parameter that are not fixed. |
|---|---|
| names | vector of character strings giving the names of the parameters (should not contain ":"). |
| ... | additional arguments to be passed from the convenience functions. |
Details
The asymptotic regression model is a three-parameter model with mean function:
$$ f(x) = c + (d-c)(1-\exp(-x/e))$$
The parameter \(c\) is the lower limit (at \(x=0\)), the parameter \(d\) is the upper limit and the parameter \(e>0\) is determining the steepness of the increase as \(x\).
Value
A list of class drcMean, containing the mean function, the self starter function,
the parameter names and other components such as derivatives and a function for calculating ED values.
Note
The functions are for use with the function drm.
See also
Examples
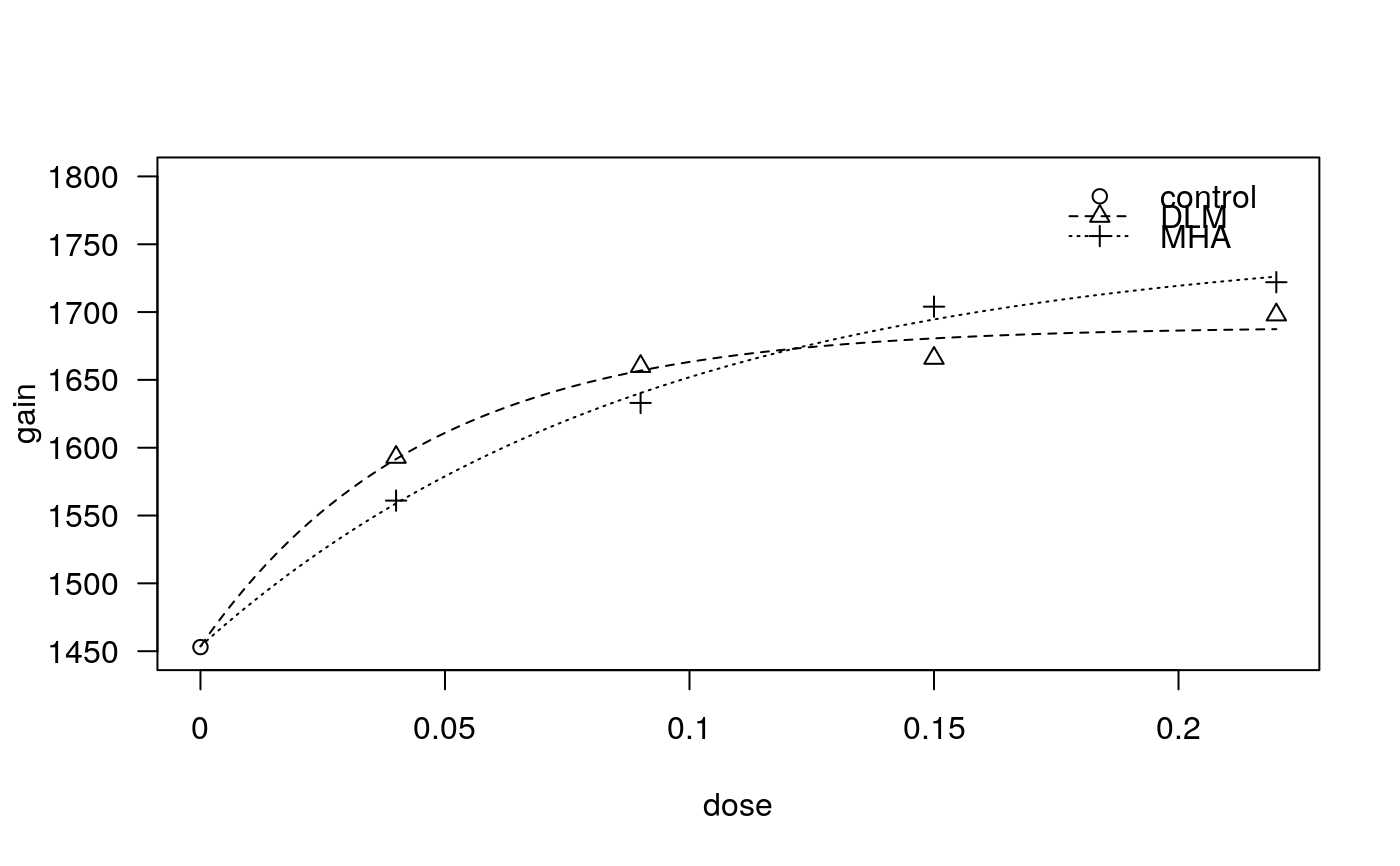
## First model met.as.m1<-drm(gain ~ dose, product, data = methionine, fct = AR.3(), pmodels = list(~1, ~factor(product), ~factor(product)))#> Control measurements detected for level: controlplot(met.as.m1, log = "", ylim = c(1450, 1800))summary(met.as.m1)#> #> Model fitted: Shifted asymptotic regression (3 parms) #> #> Parameter estimates: #> #> Estimate Std. Error t-value p-value #> c:(Intercept) 1.4536e+03 1.0764e+01 135.0395 1.804e-08 *** #> d:DLM 1.6892e+03 9.8280e+00 171.8804 6.873e-09 *** #> d:MHA 1.7541e+03 2.1369e+01 82.0855 1.320e-07 *** #> e:DLM 4.5386e-02 7.4128e-03 6.1226 0.003605 ** #> e:MHA 9.2668e-02 1.6516e-02 5.6109 0.004957 ** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Residual standard error: #> #> 11.20328 (4 degrees of freedom)## Calculating bioefficacy: approach 1 coef(met.as.m1)[5] / coef(met.as.m1)[4] * 100#> e:MHA #> 204.1797#> #> Estimated ratios of effect doses #> #> Estimate Std. Error t-value p-value #> DLM/MHA:50/50 0.4897647 0.1091974 -4.6725956 0.0094999## Simplified models met.as.m2<-drm(gain ~ dose, product, data = methionine, fct = AR.3(), pmodels = list(~1, ~1, ~factor(product)))#> Control measurements detected for level: controlanova(met.as.m2, met.as.m1) # simplification not possible#> #> 1st model #> fct: AR.3() #> pmodels: ~1, ~1, ~factor(product) #> 2nd model #> fct: AR.3() #> pmodels: ~1, ~factor(product), ~factor(product) #>#> ANOVA table #> #> ModelDf RSS Df F value p value #> 1st model 5 1861.77 #> 2nd model 4 502.05 1 10.8332 0.0302met.as.m3 <- drm(gain ~ dose, product, data = methionine, fct = AR.3(), pmodels = list(~1, ~factor(product), ~1))#> Control measurements detected for level: controlanova(met.as.m3, met.as.m1) # simplification not possible#> #> 1st model #> fct: AR.3() #> pmodels: ~1, ~factor(product), ~1 #> 2nd model #> fct: AR.3() #> pmodels: ~1, ~factor(product), ~factor(product) #>#> ANOVA table #> #> ModelDf RSS Df F value p value #> 1st model 5 1878.73 #> 2nd model 4 502.05 1 10.9683 0.0296